Our Client:
A prominent firm in the agricultural industry
Situation:
Annually, 40% of global crop production, costing the global economy over $220 billion, is lost to pests and plant diseases. This issue is expected to escalate with climate change. Recognizing the potential of an efficient crop disease detection model, the client sought to combat this growing issue.
Task:
The objective was to create a system that could accurately identify plant types and detect any present diseases using advanced machine learning techniques. This technology would serve as an augmentation to farmers' skills, enabling them to identify which crops require more nurturing and nutrients, thereby leading to a more efficient harvest season. It would also facilitate quicker and more accurate decision-making, reducing the time spent on manual inspections.
DSL implemented various machine learning models for this task, each with its own set of advantages and disadvantages. While this system might be considered niche by some, the underlying technology has broad applications in many other industries. For instance, the object detection technology used in this case study is crucial for applications such as identifying trees in forestry management or detecting animals for wildlife conservation efforts.
Result:
The initial model implemented was Faster RCNN, a robust, efficient and well understood model for object detection. However, it's an anchor-based model, which necessitates a significant amount of prior knowledge about the specific image dataset. This requirement led to an extensive exploration of the dataset, which proved to be time-consuming.
Recognizing the need for a more efficient approach, we transitioned to YOLOx, an anchor-free model. This model offered similar training times as Faster RCNN but with the added advantage of not requiring extensive dataset exploration, making it a more efficient choice.
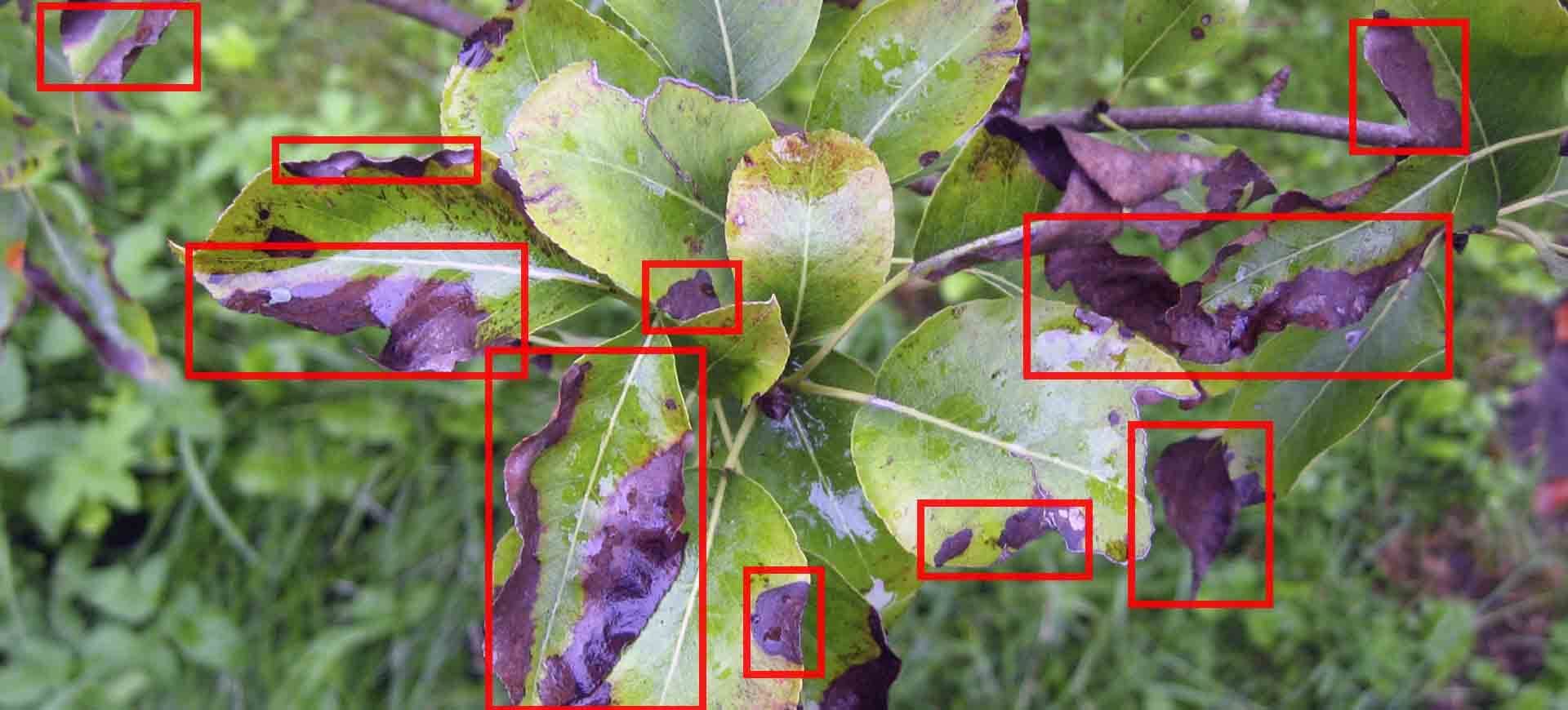
Both models, after being fine-tuned to our custom dataset, demonstrated encouraging results, achieving a commendable mAP score of 42.7, which compares favorably to the standard MS COCO baseline of 40.5. However, we faced obstacles due to the diversity of plant types and their corresponding diseases, coupled with an imbalance in the quantity of images available for each label. Some images displayed multiple plant instances, but only a few were labelled, leading to ambiguity during the learning phase. This resulted in a number of false negatives (i.e., missed detections) by our model, a consequence largely attributable to the quality of the dataset.


